Although biological monitoring using environmental DNA (eDNA) analysis is becoming increasingly popular worldwide, there is a lack of research on basic information on the characteristics (production source and persistence state) and dynamics (transport, diffusion, retention, degradation) of eDNA. We reviewed eDNA studies published in international peer-reviewed journals during 2008-2020 (728 studies in total) and indicated a particular lack of studies on their molecular and cytological states.
Panel (a) of the above figure represents the time-series changes in the number of macrobial eDNA studies. Gray scales of each bar plot show the studies corresponding to each keyword with regard to the characteristics and dynamics of eDNA, specified in this review (production process, persistence state, transport process, and degradation process from lightest to darkest), while studies not corresponding to any of the keywords are represented in white. The total number of studies published in each year is shown respectively at the top of the bar plot. Panel (b) shows the total number of studies corresponding to each keyword. The percentage (%) of the total is represented by the number in brackets respectively.
Given that the transport and degradation processes of eDNA can strongly depend on the characteristics of eDNA itself in addition to environmental factors, this result suggests a potential bottleneck regarding the basic information of eDNA. In response, we introduced some previous findings to outline the new applicability of eDNA analysis based on its various states, including nuclear DNA, longer DNA fragments, and intra-cellular DNA, compared with the current eDNA analysis based on shorter mitochondrial DNA fragments.

Author:Toshiaki S. Jo(Ryukoku University/JSPS)
Article: Jo, T., Takao, K., & Minamoto, T. (2022) Linking the state of environmental DNA to its application for biomonitoring and stock assessment: targeting mitochondrial/nuclear genes, and different DNA fragment lengths and particle sizes. Environmental DNA, 4(2), 271-283.

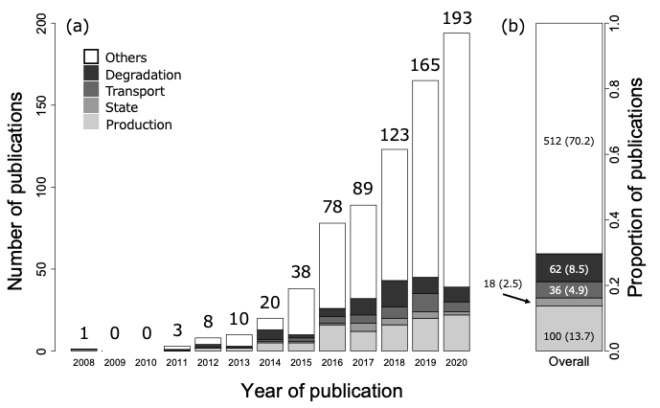 (a) Time-series changes in the number of macrobial eDNA studies. The total number of studies published in each year is shown at the top of the bar plot. (b) The total number of studies corresponding to each keyword. The percentage (%) of the total is represented by the number in brackets. [Quoting Figure 2 from the original paper]
(a) Time-series changes in the number of macrobial eDNA studies. The total number of studies published in each year is shown at the top of the bar plot. (b) The total number of studies corresponding to each keyword. The percentage (%) of the total is represented by the number in brackets. [Quoting Figure 2 from the original paper]